








Study with the several resources on Docsity

Earn points by helping other students or get them with a premium plan


Prepare for your exams
Study with the several resources on Docsity

Earn points to download
Earn points by helping other students or get them with a premium plan
Community
Ask the community for help and clear up your study doubts
Discover the best universities in your country according to Docsity users
Free resources
Download our free guides on studying techniques, anxiety management strategies, and thesis advice from Docsity tutors
this document contain the process of making RNA from DNA , known as transcription
Typology: Study notes
1 / 13

This page cannot be seen from the preview
Don't miss anything!








CORE COURSE 2: Molecular Biology Unit 3: Transcription Transcription is the process through which a DNA sequence is enzymatically copied by an RNA polymerase to produce a complementary RNA. The synthesis of RNA from a single strand of a DNA molecule in the presence of enzyme RNA polymerase is called transcription. It is the process of formation of a messenger RNA molecule using a DNA molecule. Transcription in Prokaryotes: ➢ Transcription in prokaryotes (and in eukaryotes) requires the DNA double helix to partially unwind in the region of mRNA synthesis.
➢ The region of unwinding is called a transcription bubble. ➢ Transcription always proceeds from the same DNA strand for each gene, which is called the template strand. ➢ The mRNA product is complementary to the template strand and is almost identical to the other DNA strand, called the non template strand. ➢ The only difference is that in mRNA, all of the T nucleotides are replaced with U nucleotides. In an RNA double helix, A can bind U via two hydrogen bonds, just as in A–T pairing in a DNA double helix. ➢ The nucleotide pair in the DNA double helix that corresponds to the site from which the first 5' mRNA nucleotide is transcribed is called the +1 site , or the initiation site. ➢ Nucleotides preceding the initiation site are given negative numbers and are designated upstream. Conversely, nucleotides following the initiation site are denoted with “+” numbering and are called downstream nucleotides. ➢ Upstream refers to “towards the 5’ end” and downstream refers to “towards the 3’ end”. ➢ The region of DNA that contains sequences that are the signals for transcribing a gene are termed promoters. ➢ +1 refers to the basepair where transcription starts; - x refers to x basepairs 5’ to the start site. Factors required for transcription:
4. Promoter:
❖ Tertiary complex: RNA polymerase starts synthesizing nucleotide. It doesnot require the help of primase. If the enzyme synthesize short RNA molecules of less than 10 bp, it does not further elongates which is called abortive initiation. This is because σ 3.2 acting as mimic of RNA and it lies at middle of RNA exit channel in open complex. When the RNA polymerase manage to synthesize RNA more than 10 bp long, it eject the σ 3.2 region and RNA further elongates and exit from RNA exit channel. This is the formation of tertiary complex. Summary of Initiation: RNAP scans the DNA looking for promoters
5' to 3' direction, and unwinds and rewinds the DNA as it is read.
3. Termination: There are two mechanism of termination. Rho independent: ✓ In this mechanism, transcription is terminated due to specific sequence in terminator DNA. ✓ The terminator DNA contains invert repeat which cause complimentary pairing as transcript RNA form hair pin structure. ✓ This invert repeat is followed by larger number of TTTTTTTT(~8 bp) on template DNA. ✓ The uracil appear in RNA. The load of hair pin structure is not tolerated by A=U base pair so the RNA get separated from RNA-DNA heteroduplex. ✓ RNA transcription stops when the newly synthesized RNA molecule forms a G-C rich hairpin loop, followed by a run of U’s, which makes it detached the DNA template.
Eukaryotic Transcription Eukaryotic RNA polymerase: ➢ DNA dependent RNA polymerase requires DNA as a template, four ribonucleosides 5’ triphosphate ATP, GTP, CTP and UTP as building blocks of RNA and Mg2+ as a cofactor. ➢ Eukaryotes have three different RNA polymerases specialized for transcription of different types of RNA molecules. ➢ Eukaryotic genomes are larger in the range of approximately 109 base pairs. Large genome means large number of genes and hence requires more specificity for amplification. ➢ Additionally eukaryotic cells have diversity of functions, organelles and specialized cell types etc which also require specificity of gene expressions. Hence eukaryotes have different RNA polymerases specialized for transcription of different types of RNA molecules.
➢ RNA polymerase I functions for synthesis of pre ribosomal RNA containing 18S, 5.8S and 28S rRNA. ➢ In general RNA polymerases are a large multisubunit complex consisting of 10-17 different subunits. ➢ These RNA polymerases cannot bind specifically to their respective promoters on their own but require assistance from several transcriptional factors. RNA polymerase II : ➢ RNA polymerase II is involved in synthesis of messenger RNA (mRNA). It is capable of recognizing several promoters that differ in their sequence. ➢ Many Polymerase II recognized promoters have few common sequences like the TATA box (consensus sequence TATAAA) located are – 30 position and Inr sequence (initiator) at RNA start site at +1 base pairs. ➢ RNA polymerase II is a multimeric protein made up of 12 different subunits (RBP1-12). ➢ RBP1 is the largest subunit and is homologous to β’ subunit of bacterial RNA polymerase. ➢ The RBP2 subunit is homologous to bacterial RNA polymerase β subunit, while RBP3 and RBP 11 are homologous to bacterial RNA polymerase α subunits. ➢ The largest subunit RBP1 of Pol-II has a long C terminal tail made up of multiple repeats of 7 amino acid sequence – YSPTSPS-. ➢ The RNA polymerase II requires interaction with several transcriptional factors in order to initiate eukaryotic transcription process. RNA polymerase III: ➢ RNA polymerase III is responsible for synthesizing tRNA’s, 5SrRNA and other small RNAs. Some of the promoter sequences recognized by RNA polymerase III are located within the genes while others are located upstream of the RNA start site. Eukaryotic promoter elements : ➢ Eukaryotic promoters extend from the transcriptional start site to approximately 200 bps upstream. ➢ It contains several short sequences of approximate 10 bps in length. ➢ The core promoter contain a TATA box (sequence TATAAA), which is bound by TATA binding protein that functions in the formation of the RNA polymerase transcriptional complex. ➢ The TATA box is present upstream of the transcriptional start site (-25 bps upstream) (often within 50 bases) and an INR sequence at the RNA polymerase start site. ➢ Several proximal promoter elements approximately 70-200 bps towards the 5’ end of transcription start site are also present. ➢ These includes CAAT box and GC box which are binding sites for CAAT- binding protein (CBP) and transcriptional factor SP1 respectively. ➢ These different sequences can be mixed and matched to give a functional promoter.
Fig: Eukaryotic promoter element recognized by RNA Pol II Fig: Eukaryotic promoter element recognized by RNA Pol III
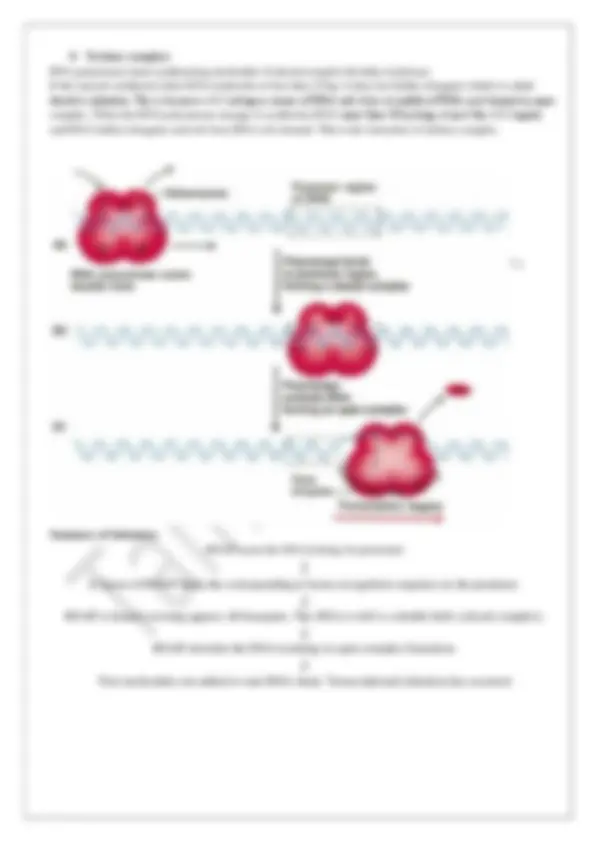
Steps of Eucaryotic Transcription: The transcription process by RNA polymerase II can be in general divided into several phases , - assembly, initiation, elongation and termination.
1. Initiation: ➢ Assembly of RNA Polymerase and Transcription Factors at a Promoter ✓ The assembly of RNA polymerase and transcriptional factors at promoter begins by formation of a closed complex which begins with the binding of TATA- binding protein (TBP) to the TATA box and 8-10 TBP associated factors. TBP is universal transcriptional factor required by all three classes of RNA polymerases. ✓ The TBP is further bound by transcriptional factor TFIIB which also binds with DNA on either side of TBP. It acts as in intermediate in the recruitment of RNA polymerase II and influences selection of transcription start site. ✓ TFIIA at this stage stabilizes TFIIB-TBP complex on the DNA. ✓ The TFIIB – TBP complex is then associated with another complex comprising of TFIIF and Pol II. ✓ TFIIF here recruits RNA polymerase II and targets it to its promoter via interaction with TFIIB and reducing nonspecific binding by RNA Polymerase II. ✓ Finally TFIIE- and TFIIH bind leading to formation of an open complex. TFIIE here acts in recruitment of TFIIH and modulates its activities. TFIIH has a helicase activity which is responsible for transition from closed to open promoter complex. ➢ RNA Strand Initiation and Promoter Clearance: ✓ Additional function of TFIIH has kinase acitivity in one of its subunits that brings about phosphorylation of Pol II at many places in its CTD. Additional protein kinase, CDK9 which is a part of transcription elongation complex pTEFb (positive transcription elongation factor b) also brings about phosphorylation of the CTD. ✓ Phosphorylation events at the CTD of RNA polymerase II largest subunit leads to a conformational change in the overall structure of the complex, initiating transcription. ✓ Phosphorylation of the CTD is important not only during the subsequent elongation phase but it also affects the interactions between the transcription complex and other enzymes involved in post transcriptional processing of the transcript. ✓ While the initial 60-70 nucleotides of RNA are being synthesized the TFIIE followed by TFIIH is released, and Pol II continues to enter into the elongation step of transcription. 2. Elongation: ✓ TFIIF is associated with Pol II throughout elongation phase. ✓ During this step, the RNA Pol II C terminal domain is maintained in the phosphorylated stage by coordinated action of several proteins called as elongation factors. ✓ The elongation factors prevent pausing of transcription process and are also involved in interaction with protein complexes that mediate post transcriptional processing of mRNA. ✓ After synthesis of the RNA molecule the process is terminated and Pol II is dephosphorylated and recycled to initiate another transcription cycle. Elongation factor Protein Function and activity P-TEFb Positive transcription elongation factor b
Fig: Transcription at RNA Pol II promoters
3. Termination: ➢ Termination by RNA polymerase I: Transcription termination by RNA polymerase I requires binding sites for Reb1p which causes pausing of transcription by RNA polymerase I and the process terminates.
Sen1-dependent termination: ✓ It is an alternative pathway for most non- coding RNAs. ✓ The 3’ ends of yeast snRNAs and snoRNAs are generated by endoribonucleolytic trimming by nuclear exosome TRAMP complex and do not possess a poly A tail in their mature form. ✓ A distinct set of core factors is required for recognition and transduction of the transcription termination signal, including the RNA-binding proteins Nrd1, nuclear polyadenylated RNA-binding protein3 (Nab3) and the putative RNA and DNA helicase Sen1. ✓ In this pathway Sen1 is proposed to terminate Pol II by unwinding the RNA–DNA hybrid in the active site.